Research
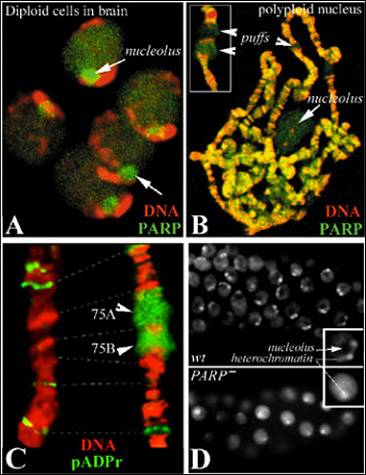
Figure 1
The principal focus of our research is on understanding functions of poly(ADP-ribose) polymerase 1 (PARP1) and the mechanisms of chromatin regulation by protein-poly(ADP-ribosyl)ation. We have recently demonstrated that poly(ADP-ribose) polymerase 1 (PARP1) and poly(ADP-ribose) glycohydrolase (PARG) perform a number of vital previously unrecognized functions in a number of cellular processes (Tulin et al. 2002; Tulin et al. 2006). Distributed evenly along chromatin (Figure 1AB) and enriched in nucleoli (Figure 1AB), PARP1 is responsible for rapid local chromatin decondensation (loosening) (Figure 1), which is required for transcriptional activation of many genes within specific chromatin blocks (Tulin & Spradling 2003). Poly(ADP-ribosyl)ation is also involved in heterochromatin formation and the initiation and maintenance of nucleoli (Figure 1D).
Publications:
Tulin A, Stewart D, and Spradling AC (2002) The Drosophila heterochromatic gene encoding
poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during
development. Genes Dev. 16:2108-2119. PubMed
Tulin A and Spradling A (2003) Chromatin loosening by poly(ADP)-ribose polymerase (PARP)
at Drosophila puff loci. Science 299:560-562.PubMed
Tulin A, Chinenov Y, and Spradling A (2003) Regulation of chromatin structure and gene activity
by poly (ADP-ribose) polymerases. Curr. Top. Dev. Biol. 56:55-83.John Hopkins University
Tulin A, Naumova N, Menon A, and Spradling A (2006) The Drosophila Poly(ADP-ribose) Glycohydrolase
(Parg) protein mediates chromatin structure and Sir2-dependent silencing. Genetics
172:363-371.PMC
H2Av Histone Controls PARP1 Protein Activation in Chromatin
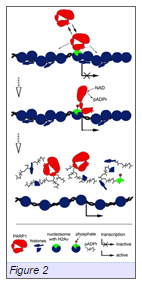
Figure2
We have successfully identified novel chromatin-associated PARP partners by implementing a Tandem Affinity Purification (TAP) strategy together with sucrose gradient purification and liquid chromatography-tandem mass spectrometry (LC-M.S./M.S.). Candidate interactors arising from this approach were analyzed functionally for their influence on PARP, using standard genetic approaches combined with immunostaining and confocal microscopy. Based on this work, the variant histone H2Av (an H2Az, H2Ax homologue in Drosophila) has been identified as a protein that promotes targeting of PARP1 to chromatin. Ser137-phosphorylated H2Av has been shown to co-localize with foci of local PARP activationin vivo(Meyer-Ficca et al., 2005). Our findings establish the importance of H2Av-PARP1 interaction in terms of associating these proteins with chromatin. We propose the following model for PARP1 binding to chromatin (Figure 2Kotova et al., 2011): (1) H2Av establishes specific distinct domains in chromatin. (2) PARP interacts with H2Av-positive chromatin. (3) Binding of PARP with chromatin is dynamic and H2Av-dependent. (4) PARP1 enzymatic activity is neither required for its initial embedding in chromatin, nor for its maintenance there. (5) Disruption of the interaction between PARP1 and H2Av by phosphorylation of H2Av or mutation in H2Av phosphorylation domain leads to spontaneous PARP1 activation.We focus our near-term efforts on validating the mechanisms of PARP1 targeting to specific chromosomal domains using an extended in vivo and in vitro analysis of PARP1-H2Av co-regulation, supported by mapping the subdomains of H2Av involved in these interactions.
Publications:
Pinnola AD, Naumova N, Shah M, andTulin AV(2007) Nucleosomal core histones mediate dynamic regulation of PARP1 protein binding
to chromatin and induction of PARP1 enzymatic activity. J. Biol. Chem. 282: 32511-32519.PubMed
Kotova E, Jarnik M, andTulin AV(2009) Poly (ADP-ribose) Polymerase 1 is required for protein localization to Cajal
body. PLoS Genetics. 5(2): e1000387.PubMed
Kotova E, Jarnik M, andTulin AV(2010) Uncoupling of the trans-activation and trans-repression functions of PARP1
protein. Proc. Natl. Acad. Sci. USA. 107(14): 6406-6411.PubMed
Kotova E, Lodhi N, Jarnik M, Pinnola AD, Ji Y, andTulin AV(2011) Drosophila histone H2A variant (H2Av) controls Poly(ADP-Ribose) Polymerase
1 (PARP1) activation in chromatin. Proc. Natl. Acad. Sci. USA. 108(15): 6205-6210.PubMed
Thomas CJ, Kotova E, Andrake M, Adolf-Bryfogle J, Glaser R, Regnard C, andTulin AV(2014) Kinase-mediated changes in nucleosome conformation trigger chromatin decondensation
via poly-ADP-ribosylation. Molecular Cell. 53: 831–842. PubMed
Poly(ADP-Ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates gene expression
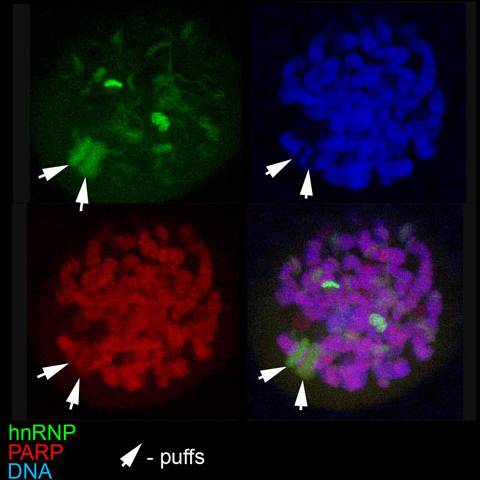
Within the short span of the cell cycle, Poly(ADP-ribose) (pADPr) can be rapidly produced by Poly(ADP-ribose) Polymerases (PARPs) and degraded just as rapidly by Poly(ADP-ribose)Glycohydrolases (PARGs). The biological significance of pADPr oscillations in a living cell in steady state conditions remains unclear. Recently, we identified heterogeneous nuclear RNP-binding proteins (Hrps) as preferential PARP targets, which mediate PARP1 regulation of gene expression during development (Figure 3Ji & Tulin 2009). Here, we focus on investigating how changes in poly(ADP-ribosyl)ation of Hrp38 regulate tissue specification and planar polarity during oogenesis in Drosophila. Specifically, turnover of poly(ADP-ribose) regulates Drosophila germ-line stem cell maintenance and oocyte positioning by modifying Hrp38 protein function. Hrp38 binds to the 5’UTR of DE-cadherin to promote its translation. Furthermore, pADPr binding to Hrp38 in germ-line stem cell progeny disrupts the interaction of Hrp38 with the 5’UTR of DE-cadherin mRNA, thereby reducing DE-cadherin translation. Blocking the translation of DE-cadherin leads to its depletion, which, in turn, causes differentiation of progenitor cells. In agreement with these results, we found that defects in either pADPr catabolism or Hrp38 function cause a decreased expression of DE-cadherin, which leads to a loss of germ-line stem cells and mislocalization of oocytes in the ovary. Taken together, our findings suggest that Hrp38 and its poly(ADP-ribosyl)ation regulate DE-cadherin expression, and by doing so, control germ-line stem cell maintenance and oocyte localization on the level of translation. Since PARP utilizes cellular NAD to produce pADPr and its activity ultimately depends on the energy status of an organism, we establish the chain of events that connect extrinsic conditions with the mode of development and functioning of an organism.
Publications:
Ji Y andTulin AV(2009) Poly(ADP-ribosyl)ation of Heterogeneous Nuclear Ribonucleoproteins Modulates
Splicing. Nucl. Acids Research. 37: 3501-3513.PubMed
Ji Y andTulin AV(2010) The roles of PARP1 in gene control and cell differentiation. Review for Curr.
Opin. Genet. Dev. 20(5):512-518.PubMed
Ji Y andTulin AV(2012) Poly(ADP-ribose) controls DE-cadherin-dependent stem cell maintenance and oocyte
localization. Nat Communications. 3:760. doi:10.1038/ncomms1759.PubMed
Ji Y andTulin AV(2013) Post-transcriptional regulation by Poly(ADP-ribosyl)ation. Int J Mol Sci. 14:
16168-16183.PMC
Ji Y andTulin AV(2013) Alternative Splicing Regulation by Poly(ADP-ribosyl)ation. Nova Science Publishers.
New Development in Alternative Splicing Research. pp. 159-170.
Ji Y, Jarnik M, andTulin AV(2013) Poly(ADP-ribose) Glycohydrolase and Poly(ADP-ribose)-interacting Protein Hrp38
Regulate Pattern Formation during Drosophila Eye Development. Gene. 526: 187–194.PubMed
Ji Y, Thomas C,Tulin N, Lodhi N, Boamah E, Kolenko V, and Tulin AV (2016) Charon mediates IMD-driven PARP-1
dependent immune responses in Drosophila. Journal of Immunology. 197: 2382-2389.PMC
Ji Y andTulin AV(2016) Poly(ADP-ribosyl)ation of hnRNP A1 protein controls translational repression
in Drosophila. Molecular and Cellular Biology. 36(19): 2476-2486.MBC
Poly(ADP-Ribose) Polymerase 1 Regulates Nucleolar Functions and Translation
Poly(ADP-ribose)polymerase 1 (PARP1) is a nuclear protein that utilizes NAD to synthesize poly(ADP)ribose (pADPr), resulting in both automodification and the modification of acceptor proteins. Substantial amounts of nuclear PARP1 and pADPr (up to 50 %) (Figure 1) are localized in the nucleolus, a subnuclear organelle where ribosomal biogenesis and maturation occur (Tulin et al., 2002, Tulin et al., 2006). At present, the functional significance of PARP1 protein inside the nucleolus remains unclear. In this project, we investigate the roles that PARP1, pADPr, and PARP1-interacting proteins play in the maintenance of nucleolus structure and functions. Our analysis shows that PARP1 interacts with a select group of nucleolar proteins. Moreover, PARP1 is required for targeting these proteins to the proximity of pre-rRNA; hence, PARP1 controls pre-rRNA processing, post-transcriptional modification and pre-ribosome assembly. Importantly, we have observed that disruption of PARP1 enzymatic activity causes nucleolus disintegration and aberrant localization of nucleolar-specific proteins. Based on these findings, we propose a model that explains how PARP1 activity impacts nucleolar functions and, consequently, ribosomal biogenesis.
During our preliminary research, we have identified a subset of ribosomal proteins that bind preferentially to PARP1 and potential PARP1 targets, and, therefore, may influence PARP1 regulation of nucleolar functions and translation during development and stress response (Pinnola et al., 2007). Previous studies by other research groups have confirmed strong PARP1 interaction with ribosomal protein L22 (rpl22) and ribosomal protein L23 (rpl23) in vivo (Koyama et al., 1999). Subsequently, rpl22 protein was also suggested to control global IRS-dependent translational initiation (Dr. David Wiest and Dr. Randy Strich, personal communication). We are currently investigating PARP1 regulation of translation and nucleolar integrity. We began by performing a functional study of the genetic and biochemical interaction of PARP1 with rpl22 and RPL23. In addition, we plan to investigate the mechanisms by which PARP1 regulates nucleolar functions and translation.
Publications:
Boamah EK, Kotova E, Garabedian M, Jarnik M, and Tulin AV (2012) Poly(ADP-ribose) Polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genetics. 8(1):e1002442. PubMed
PARP1 and pADPr are Epigenetic Marks in Mitotic Chromatin
The goal of this project is to determine how different kinds of new epigenetic marks are distributed and stabilized specifically in mitotic chromatin. While many transcription factors, co-regulators, and histone modifications exist in the interphase chromatin and are important for gene regulation, those which occur in mitotic chromatin can clearly confer nonmutational, epigenetic inheritance. Histone modifications are broadly associated with either active or inactive genes, but do not set or re-set precise gene expression states. Such states can be set by transcription factors and co-regulators. However, most those that have been investigated, do not occupy mitotic chromatin and hence cannot function as epigenetic marks. We have recently discovered that Poly(ADP-ribose) Polymerase 1 (PARP1), which loosens local chromatin, along with poly(ADP ribose) (pADPr) itself, are distinct from many other factors in that they stably occupy mitotic chromatin. Given the ability of these factors to remain bound to chromatin through mitosis and their ability to establish a transcriptionally competent state, we propose that PARP1 and pADPr, serve as a new class of epigenetic marks. We are examining the role of poly-ADP-ribosylation in epigenetic regulation. Specifically, we plan to identify sites of Poly(ADP-ribose) Polymerase 1 (PARP1) binding to mitotic and interphase chromatin. We will also purify proteins associated with mitotic chromatin to uncover new epigenetic regulatory factors. Our focus on mitotic events and new classes of epigenetic marks provide novel directions for the field. Our work will reveal a mechanistic basis of epigenetic inheritance and hence functional targets for developing therapeutic drugs to ultimately treat epigenetics-based human diseases.
Publications:
Lohdi N and Tulin AV (2011) PARP1 Genomics: Chromatin Immunoprecipitation Approach using Anti-PARP1 Antibody
(ChIP and ChIP-seq). Methods Mol Biol. 780: 191-208.
Lodhi N, Ji. Y, and Tulin AV (2016) Mitotic bookmarking: maintaining post-mitotic reprogramming of transcription
reactivation. Curr Mol Biol Rep. 2:10-16.
Lodhi N, Kossenkov A, and Tulin AV (2014) Bookmarking promoters in mitotic chromatin: Poly(ADP-ribose)Polymerase-1 as
an epigenetic mark. Nucleic Acids Res. 42(11):7028-7038. PubMed
Poly(ADP-Ribosyl)ating Enzymes in Aging Control
In a eukaryotic organism, the progression of aging is associated with molecular changes that regulate the transcriptional activation of age-related genes, the capacity for DNA repair, programmed cell death, and a number of other vital processes. Accelerated by environmental stress and damage accumulation, such changes ultimately lead to an exponential increase in frailty and morbidity, and ultimately control the lifespan of an organism. Poly(ADP-ribosyl)ation of proteins has long been known as a posttranslational modification that is coupled to DNA repair and apoptosis triggering. Recently, the crucial role of PARPs in a number of developmentally regulated processes, including chromatin remodeling, transcriptional regulation, and telomere elongation has also been recognized.
We plan to investigate how the regulation of a single pathway, the turnover of poly(ADP-ribose) (pADPr), achieves the linkage between transcriptional regulation of specific genes on the one hand and organismic aging and longevity on the other. Our preliminary results show that PARP1 activity is significantly elevated in aging Drosophila. A considerable extension of the lifespan was observed for a fly strain that has only one copy of the PARP1 gene when compared with wild type flies that carry two copies of this gene. We have also demonstrated that PARG/PARP1 controls the activation of a number of age specific genes. Knockdown of these genes leads to a pronounced extension of life, whereas ectopic expression of these genes in young flies causes premature aging and death. Additionally, we found that degree of poly(ADP-ribosyla)ation of Lamin C--a protein involved in transcriptional silencing--is increased in old flies. These findings allow us to propose that PARP1 protein is a principal regulator of the aging program in Drosophila that controls aging program via regulation of transcription of age associated genes.
New Approach to Identifying Poly(ADP-Ribose) Polymerase Inhibitors
During the past few years, Poly(ADP-ribose)Polymerase (PARP) proteins have become a popular target for anti-cancer treatment. Many PARP inhibitors have been generated and tested by the pharmacological industry. However, most of these inhibitors were designed to disrupt the DNA-dependent PARP1 protein activation pathway, based on competition with NAD for a binding site on the PARP molecule.This limitation resulted in discovery of nucleotide-like PARP1-inhibitors that may target not only PARPs, but also other enzymatic pathways involving NAD and nucleotides as co-factors. We are exploring a strategy for the identification of PARP inhibitors that target a different pathway, histone-dependent PARP1 activation. In addition to identification of NAD competitors in a small molecules collection, this approach allows the discovery of novel classes of PARP inhibitors that only disrupt histone-based steps of PARP1 activation and therefore are more specific and more effective. This project will result in development of novel small molecule inhibitors, which in future could be applied to anti-cancer treatment.
Publications:
Kotova E, Pinnola AD, and Tulin AV (2011) Small-molecule Collection and High-throughput Colorimetric Assay to Identify
PARP-1 Inhibitors. Methods Mol Biol. 780: 491-516 PubMed
Tulin A (2011) Re-evaluating PARP1 inhibitor in cancer. Nat Biotechnol. 29(12):1078-9. Nature Biochemistry
Kirsanov KI, Kotova E, Markov P, Golovine K, Lesovaya EA, Kolenko V, Yakubovskaya
MG, Tulin AV(2014) Minor grove binding ligands disrupt PARP-1 activation pathways. Oncotarget.
5: 428-437. PMC
Thomas C, Ji Y, Kotova E, Lodhi N, Pinnola AD, Golovine K, Makhov P, Pechenkina K,
Kolenko V, Tulin AV (2016) New generation non-NAD-like PARP-1 inhibitors effectively eliminate drug resistant
tumors in vivo. EBioMedicine. 13 (2016) 90–98. Science Direct