Ongoing Projects
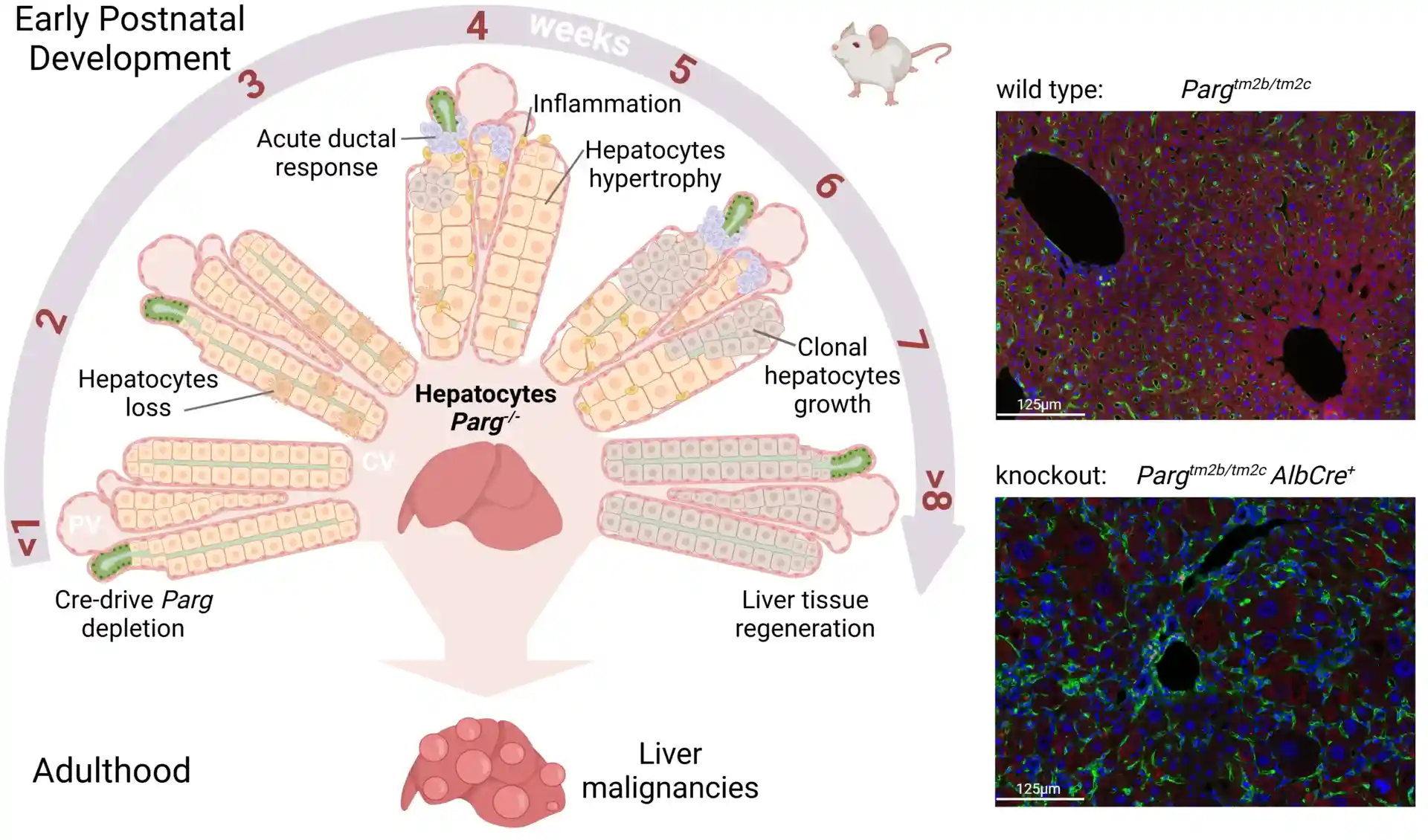
Takling Hepatocellular Carcinoma
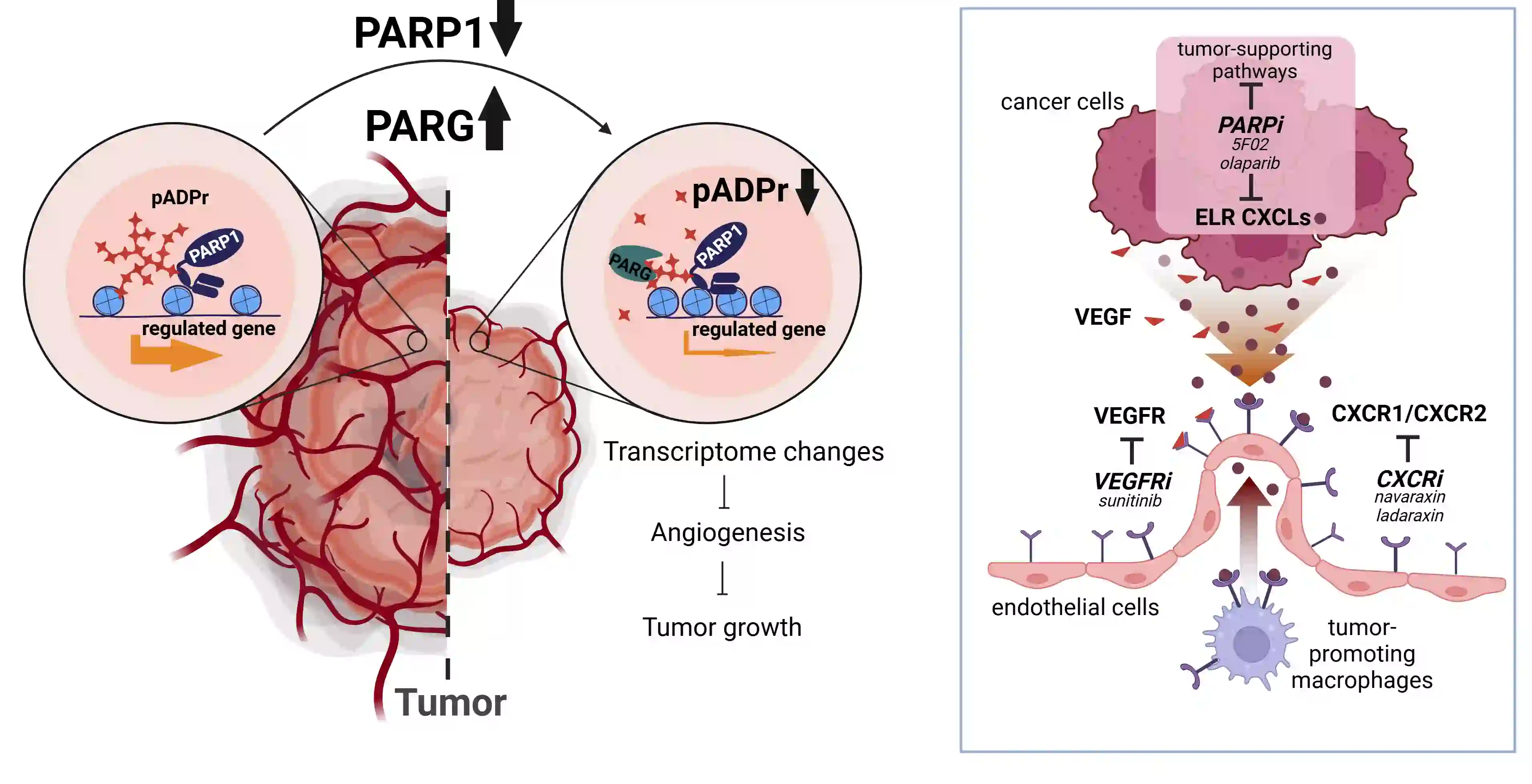
Poly ADP ribose polymerase (PARP) is an abundant ubiquitous NAD-dependent nuclear enzyme that mediates important steps of DNA repair, transcription and apoptosis. The majority of the molecules of this protein remains enzymatically inactive and undergoes bursts of activation for limited periods of time until they get automodified and revert to an inactive state. PARG (Poly ADP ribose polymerase glycohydrolase) is an enzyme that degrades PARP by perpetually cleaving poly ADP ribose (PAR). The poly ADP ribosylation pathway is almost always disrupted in cancer. Our preliminary data indicate that levels of PAR are increased in prostate cancer and renal cell carcinoma (RCC). Using western blotting, different cell-based assays, and quantitative, we are investigating how different proteins of the poly ADP ribosylation pathway are expressed and regulated in vitro in androgen receptor-independent cell line (PC3), primary RCC, and breast cancer. Our research team is using a Lenti-X Tet-one inducible expression system for overexpressing PARG in cancer cell lines of our interest. We aim to find out the effect of PARG overexpression in vivo (xenograft) in tumorigenesis. Cancer progression requires either the depletion of PARG or high levels of PARP-1. Therefore, genetic removal of PARP-1 or over-expression of PARG may suppress various tumor-promoting genes modulated by PARP-1. To test whether overexpression of PARG can affect tumor-promoting genes modulated by PARP-1, Lenti-X Tet-One inducible expression system (Clontech) is used to create a human PARG overexpression system. Lentiviral construct pLVX-TetOne-hPARG-Puro expresses hPARG under the regulation of Tet-On promoter, so it is expressed hPARG only in the presence of doxycycline. Human prostate, kidney, and breast cancer cell lines are transduced with Tet-One lentiviruses, the single clone which has the highest expression level of hPARG is selected and established into a stable expression cell line. The effects of overexpression of hPARG on tumor-promoting genes modulated by PARP-1 are analyzed by Western blotting, real-time PCR and RNA sequence. Currently, we are using cellular in vitro assays and high throughput imaging modalities to test several compounds for inhibitory activity against the chromatin regulating proteins PARP-1 and PARG. Examples of assays include cell migration and colony formation. In parallel, we are utilizing molecular approaches to determine the functional effects of overexpression and downregulation of the PARP-1/PARG axis in cancer. Ultimately, we plan to translate lead compounds into PDX models for testing therapeutic efficacy and safety of these inhibitors in vivo.
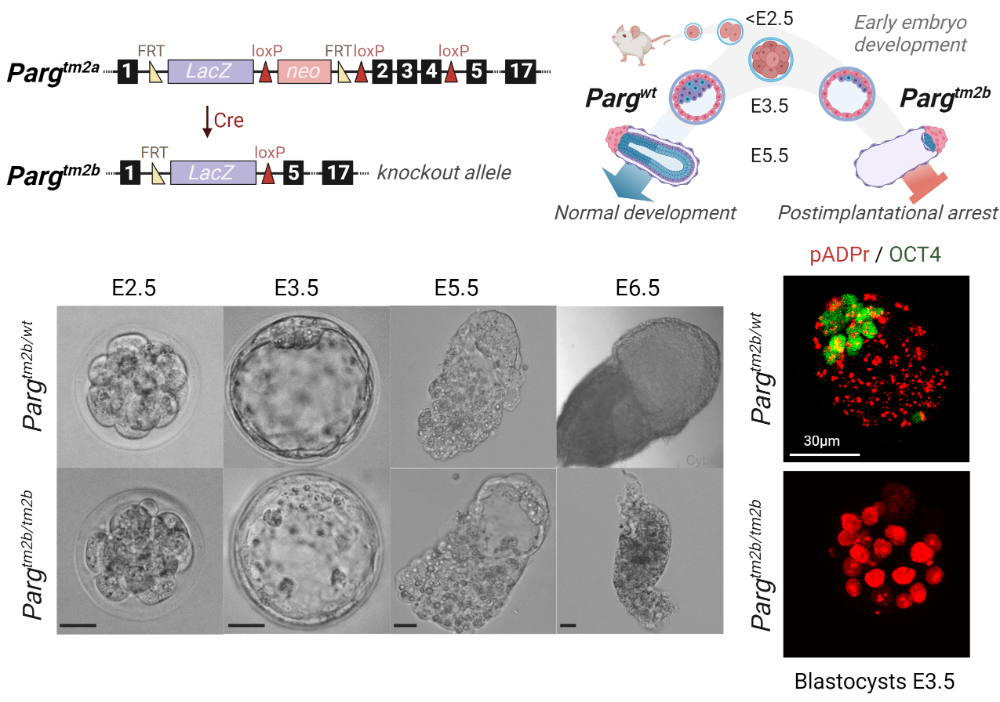
PARG in Mammalian Development
The main goal of this project is to study PARG functioning in mammals. PARG is the only enzyme that can effectively hydrolyze poly(ADP-ribose) (pADPr) chain, synthesized by different members of PARP family. PARG regulates pADPr turnover and participates in processes that require poly(ADP-ribosyl)ation. It plays a vital role in gene activation, chromatin remodeling, and regulates the maintenance of nuclear architecture. But the direct research on PARG involvement in regulating different pathways is very limited. In this project we are focusing on PARG’s role in the embryonic development. We use two mice models: a complete PARG knockout and a conditional knockout, for which the knockout can be induced during different developmental stages. Mice that completely lack PARG become arrested early in their embryonic development; we are studying factors that cause such extreme lethality. The conditional model allows as to overcome this limitation and investigate PARG role during the later stages of development. For in vitro study of cell differentiation, we use mouse embryonic stem cells, combining molecular methods of genes editing with immunohistochemistry, RNA-seq, and CHIP-seq analysis.
PARP-1 in the Transcriptional Control
PARP-1 is a multidomain nuclear enzyme. Its functions include DNA repair, chromatin remodeling, and transcriptional regulation. It contains three major binding domains. The N-terminal DNA binding domain consists of three Zinc fingers. The middle automodification domain interacts with the same domain on another PARP-1, to form PARP-1 dimers. The C-terminal catalytic domain consists of the protein interacting WGR motif and the NAD interacting site for PARP-1 (Thomas et al., 2019). PARP-1 inhibition has been used to treat cancer, inflammation, circulatory shock, stroke, and myocardial infarction. Classical PARP-1 inhibition uses NAD mimetics. However, several other cellular pathways also require NAD as a substrate leading to deleterious off-target effects from these types of inhibitors. Elucidating the mechanisms of PARP-1 binding domains will help us to create more specific and effective treatments. PARP-1 has 17 paralogs in the human genome. The Drosophila genome contains only one PARP gene, making it the perfect organism for our study. We have created 12 deletional isoforms for each binding domain of PARP in Drosophila. Each isoform was tagged with a YFP sequence. We propose to perform ChIP-seq analysis for each deletional isoform of PARP to determine which genes and cellular processes are regulated by each of PARP’s binding domains.
During tissue differentiation, gene expression programs are fixed by Polycomb and Trithorax groups (PcG and TrxG), which mark chromatin with repressive or active histone modifications that recruit effector proteins to establish local chromatin states. We identify PARP1 and PARG as critical effectors of TrxG and PcG, respectively, showing that PARP1 binds TrxG-generated histone marks, drives poly(ADP-ribose) synthesis to maintain open chromatin, while PARG associates with PcG loci to support repression.
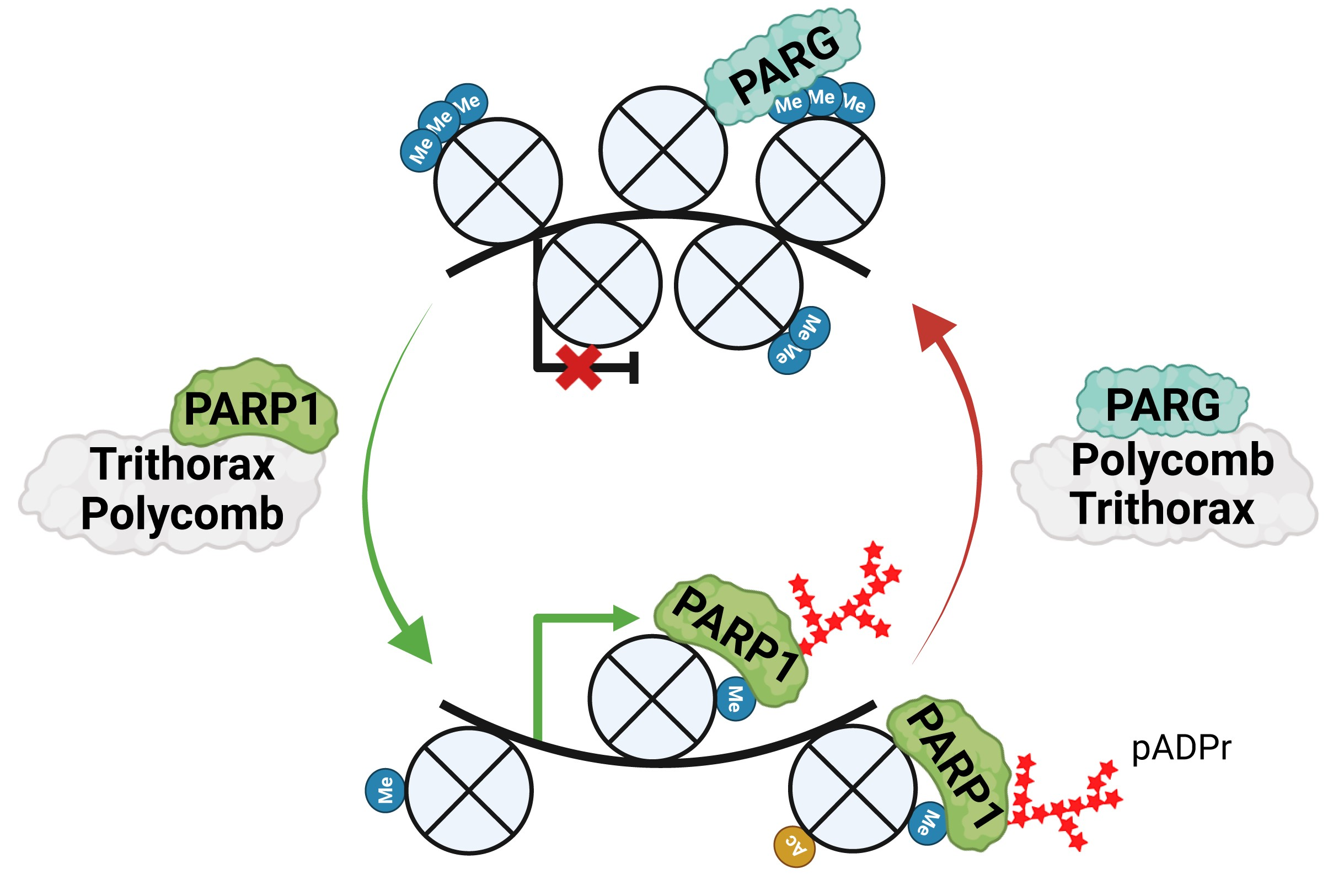
Synergy Between PARP-1, PARG Represses Transcription
Poly(ADP-ribose) polymerase 1 (PARP-1) and poly(ADP-ribose) glycohydrolase (PARG) are well known antagonistic regulators of the poly(ADP-ribose) (pADPr) metabolism, as PARP-1 assembles pADPr and PARG degrades it. The accumulation and degradation of pADPr is involved in several nuclear processes, including the regulation of chromatin structure and gene expression (Ji et al. 2016). Aberrations in pADPr metabolism have been linked to carcinogenic transformations and progression of many types of malignant tumors, as well as the development of several neurodegenerative diseases (Thomas et al 2016). Despite the clinical relevance of the pADPr metabolism, the mechanisms coordinating PARP-1 and PARG activities in pADPr metabolism remain poorly understood. By observing nucleus of wild type third instar larvae salivary gland cells, our research team has shown that a small fraction of PARG co-localizes with chromatin (Figure 1 upper panels) (Kotova et al. 2009). This fraction seems to co-localize with PARP-1 as well suggesting that they can sit together at chromatin. We found that in absence of PARG, PARP-1 fails to co-localize with chromatin and is miss located in cajal body (CB) suggesting that the presence of PARG is essential for the correct localization of PARP-1. Taken together, these results suggest that despite their antagonistic role PARP-1 and PARG can cooperate. Several lines of evidence suggest that they are involved together in the repression of the expression of several genes. To study this, we developed several line of experiment. First, to identify PARG binding sites in the DNA, we are investigating the PARG/chromatin interaction using ChIP-seq. Second, by comparing their binding profile with the expression profile of every gene in a PARG or PARP-1 mutant background, we will identify a molecular mechanism of PARP-1/PARG co-regulation. Third, by generating different protein versions of PARG, we are investigating the molecular mechanism of PARG in gene repression to bring out the PARG domains that are essential for the PARP-1/PARG synergy. Finally, we will identify the co-regulators of the PARP-1/PARG pathway in gene repression, by testing whether knockdowns of the candidate genes affect the proper function of these enzymes.