Tulin Lab
Overview of Research Interests
Chromatin Programming and Transcription
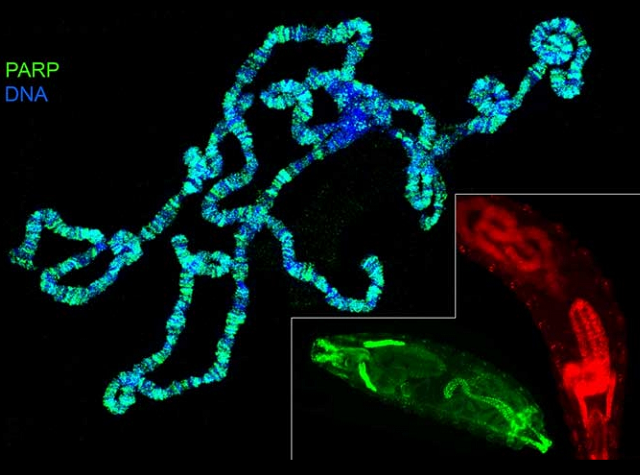
Developmentally regulated gene expression in multicellular eukaryotes requires the formation of dynamic tissue-specific chromatin structures that participate in activating certain genes and silencing others (Wolffe, Essays Biochem. 37:45, 2001). Furthermore, during an organism’s lifetime the established pattern of chromatin needs to be quickly reprogrammed in a response to environmental or hormonal signals (Thummel, Insect Biochem. Mol. Biol. 32:113, 2002). It has become increasingly clear that key aspects of chromatin structure and gene transcriptional activity are determined by a specific modification of histones (Elgin and Workman, Curr Opin Genet Dev. 12: 127, 2002; Fischle et al., Nature. 425:475, 2003).
Despite this exciting progress there remains much to be learned about how chromatin
is programmed and how active or inactive domains are maintained. Our studies of the abundant nuclear enzyme poly(ADP-ribose)
polymerase (PARP) and poly(ADP- ribose) glycohydrolase (PARG) demonstrated that it
plays novel and previously unknown roles in many of these processes. Distributed
evenly along chromatin PARP is responsible for rapid local chromatin decondensation
(loosening), which is required for transcriptional activation of many genes within
particular chromatin blocks. Previously we have demonstrated that poly(ADP-ribosyl)ation
is also involved in heterochromatin formation, the initiation and maintenance of
nucleoli and telomere metabolism. The presence of several PARP-related proteins in
mammals complicates the analysis and interpretation of results. Fortunately, only
a single PARP gene is present in the Drosophila genome, making this animal an invaluable
model system to study PARP function. Using Drosophila we study the molecular mechanisms
of PARP activation, its action on chromatin and the interaction of PARP with other
components of the chromatin remodeling machinery and transcriptional apparatus.